Medicine:Severe intellectual disability-progressive spastic diplegia syndrome
| Severe intellectual disability-progressive spastic diplegia syndrome | |
|---|---|
| Other names | CTNNB1 syndrome, CTNNB1-related intellectual disability, Intellectual disability, autosomal dominant 19[1] |
 | |
| Specialty | Medical genetics, Pediatry |
| Causes | Genetic mutation |
| Frequency | Rare, only 1 out of 50,000 live births have this condition |
Severe intellectual disability-progressive spastic diplegia syndrome is a rare novel genetic disorder characterized by severe intellectual disabilities, ataxia, craniofacial dysmorphisms, and muscle spasticity.[2] It is a type of autosomal dominant syndromic intellectual disability.[3][4][5][6][7]
Signs and symptoms
Individuals with this condition typically show severe intellectual disability, motor delays, severe speech delay and difficulties, infancy-onset hypotonia affecting the trunk, progressive hypertonia affecting the distal limbs, severe progressive microcephaly, autistic-like symptoms, aggressive behavior towards others and/or oneself, sleep abnormalities, and mild facial dysmorphisms such as a broad nose, hypoplastic alae nasi, an elongated/flattened philtrum, and a thin upper lip.[8][9]
Other symptoms include seizures, nearsightedness, farsightedness, strabismus, syringomyelia, ventriculomegaly, corpus callosum hypoplasia, hearing difficulties, and a delay in CNS myelination.[10]
In rarer cases, a child with the condition might be born with polydactyly.[11]
Complications
Most symptoms which characterize this condition arise as a result of the profound intellectual disability associated with this condition, these symptoms can cause more complications, such as communication difficulties, poor school performance, etc.[8]
Diagnosis
This condition can be diagnosed through physical examination and genetic testing.
Genetics
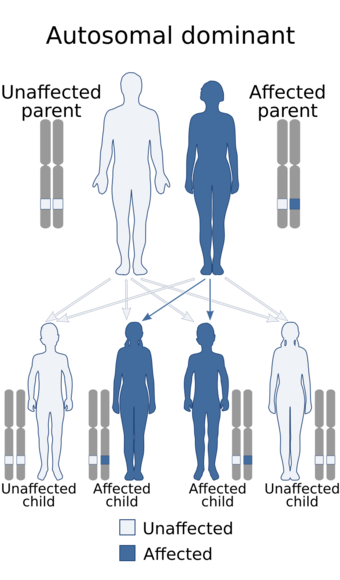
This condition is caused by heterozygous mutations in the CTNNB1 gene, located in the short arm of chromosome 3.[12][13][14][4][15] The condition, although genetic, is typically not inherited, except in rare cases where it is,[16] as it is usually the result of a de novo mutation.[17] Insertions, deletions, and other types of mutations have been reported.[18]
This gene produces a protein called "beta-catenin", which is present in various cells and tissues within the body. It is important for cell adhesion, cell communication, cell signaling, and in the normal development and function of hair follicles.[19][20][21]
Treatment
Treatment methods include occupational therapy, physical therapy, speech therapy, assistive services, constant monitoring and family counseling.[22][23]
Prevalence
This condition affects 1 out of 50,000 children worldwide.[24]
Cases
The following list comprises some cases of severe intellectual disability-progressive spastic diplegia syndrome (from the OMIM page for this condition):
- 2012: de Ligt et al. describes 2 patients from a group of 100 patients with severe intellectual disability, said 2 patients had a mutation in the CTNNB1 gene. One of these patients was a 29-year-old woman (named patient 70) who started showing signs of developmental regression when she was 6 months old, which was then followed by a moderate-severe developmental delay. She hit the milestone of sitting at the age of 2 years old and learned how to walk at the age of 12. Her first words were spoken between the ages of 9 and 10. She started showing behavioral anomalies at a young age, which consisted in aggressive behavior, self-harming behavior by automutilation, and her recurrent acts of smearing fecal matter on nearby surfaces. She was affected by microcephaly and short stature at the age of 29. She also had other mild facial dysmorphisms, such as high-arched, narrow palate, posteriorly rotated small ears, long philtrum, and low columella. They also found another patient with CTNNB1 mutation out of a group of 785 patients with intellectual disability. All of these patients had severe intellectual disabilities, speech delays, microcephaly, spasticity, and walking difficulties.[25]
- 2014: Tucci et al. describes a fourth affected patient. The patient had microcephaly, thin upper lip, autism, early-onset hypotonia, progressive spastic diplegia, and hypoplasia of the corpus callosum.[26]
- 2014: Dubruc et al. describes a 5 and a half year old girl of Caucasian descent born to healthy, un-related parents. She was affected by fetal/antenatal growth delays. Her birth was eventful, as her umbilical cord ended up wrapped around her neck and consequently, in neonatal respiratory distress syndrome. She started showing symptoms such as progressive microcephaly, hypotonia of the trunk, psychomotor delays, and speech impairments. She had a friendly and energetic personality, the latter of which was due to her hyperactivity. She had fair skin, sparse, thin hair, low-set ears, ataxia, spasticity, hyperreflexia, thin lips, a sacral dimple, and was farsighted. Genetic testing found a heterozygous 333-kb deletion in chromosome 3 which deleted the entire CTNNB1 gene and the last 3 exons of the ULK4 gene, his deleterious mutation was not found in both of her parents, thus leaving with the conclusion that it was sporadic, and not inherited.[27]
- 2015: Kuechler et al. describes 16 patients from 15 families who were confirmed to have the disorder through genetic testing, there was an equal distribution of affected males and affected females (8 girls and 8 boys). The following paragraph comprises the symptoms, with the ones listed at the top being the most prevalent ones and the ones listed at the bottom being the least prevalent ones; intellectual disabilities, infancy-onset psychomotor delays, speech impairments, facial dysmorphisms, spasticity, visual refractory errors, autistic-like symptoms with aggression, and microcephaly. Seizures and hearing impairments were not observed.[28]
- 2017: Kharbanda et al. describes 11 patients with the condition identified through interrogation of the DECIPHER database. Said patients had a mutation of the inactivating kind in the CTNNB1 gene. Out of the 11 patients, 10 had available medical details, these included intellectual disabilities, progressive microcephaly, hypotonia of the trunk, peripheral spasticity, behavioral problems, mild facial dysmorphisms, fair skin and hair, and abnormal hair patterns.[29]
- 2017: Li et al. describes a 1-year, 3 month old boy of Chinese descent who had retinal detachment and vitreous and lens opacity. He had no reaction to light. After testing negative for the mutations involved in exudative vitreoretinopathy, Li et al. found a heterozygous de novo nonsense mutation in his CTNNB1 gene. Other features exhibited by the child included developmental delays, mild thumb adduction, and microcephaly.[30]
- 2017: Panagiotou et al. describes a 3-year-old boy of Chinese descent with exudative vitreoretinopathy, developmental delays, and facial dysmorphisms, who was later found to have a de novo 1 bp insertion in his CTNNB1 gene.[31]
Animal model
In 2004, Tucci et al.[full citation needed] made an animal model consisting of mice (named "batface") with a heterozygous mutation (later named "T653K") located in the C-terminal amardillo repeat of the CTNNB1 gene. Said mice exhibited craniofacial dysmorphisms such as short nose, broad face, short anteroposterior axis. Altered brain morphology with signs such as larger structures of the deep brain, reduced volume of the cerebellum and the olfactory bulb, and corpus callosum hypoplasia. Altered behavior and cognitive function with signs such as motor defects, less complexity in vocalization, poor hippocampus-dependant memory, and deficits in pre+pulse inhibition. In vitro studies revealed that the T653K mutation 'disrupted the association between CTNNB1 and cadherin', which was consistent with a "dominant-negative effect". Abnormally high length and number of neurons, alongside diminished dendritic branching was present in the brains of heterozygous mutant mice. CTNNB1 knockdown (by the usage of siRNA) resulted in decreasing of neuronal processes and length, which lead the researchers to believe T653K is a loss-of-function type of mutation. Electrophysiologic tests showed that the neurons of mutated mice exhibited higher neural network excitability alongside decreased efficiency of functional connectivity. These results showed that CTNNB1 is important in various aspects of neurodevelopmental and synaptic function.[26]
Organizations
The following list comprises some of the organizations which help patients with this condition:
The CTNNB1 Foundation is an organization for the parents of children with the condition whose main goal is to be able to give children with CTNNB1 syndrome the opportunity of a gene therapy treatment.[32]
The CTNNB1 Syndrome Awareness Worldwide is an organization which aims at increasing awareness of this condition across the world. It also provides information on CTNNB1 syndrome to parents of children with the condition.[33]
The CureCTNNB1 is an organization which helps raise funding for research on CTNNB1 syndrome.[34]
Media coverage
This condition has been covered by a small number of news networks from Spain .[35][36][37][38][39]
See also
- FBXW7 neurodevelopmental disorder
- CHAMP1-associated intellectual disability syndrome
References
- ↑ "NIH GARD Information: Severe intellectual disability-progressive spastic diplegia syndrome". U.S. National Institutes of Health (NIH) Genetic and Rare Diseases Information Center (GARD). 16 June 2022. https://rarediseases.org/gard-rare-disease/severe-intellectual-disability-progressive-spastic-diplegia-syndrome/.
- ↑ "Severe intellectual disability-progressive spastic diplegia syndrome (Concept Id: C3554449)". MedGen - National Center for Biotechnology Information (NCBI). U.S. National Library of Medicine. https://www.ncbi.nlm.nih.gov/medgen/767363.
- ↑ "A de novo CTNNB1 nonsense mutation associated with syndromic atypical hyperekplexia, microcephaly and intellectual disability: a case report". BMC Neurology 16 (1): 35. March 2016. doi:10.1186/s12883-016-0554-y. PMID 26968164.
- ↑ 4.0 4.1 "A de novo CTNNB1 Novel Splice Variant in an Adult Female with Severe Intellectual Disability" (in English). International Medical Case Reports Journal 13: 487–492. 2020-10-07. doi:10.2147/IMCRJ.S270487. PMID 33116939.
- ↑ "KEGG DISEASE: Autosomal dominant intellectual developmental disorder". https://www.genome.jp/dbget-bin/www_bget?H00773.
- ↑ "autosomal dominant intellectual developmental disorder 19 Disease Ontology Browser - DOID:0070049". http://www.informatics.jax.org/disease/DOID:0070049.
- ↑ "Open Targets Platform" (in en). https://platform.opentargets.org/.
- ↑ 8.0 8.1 INSERM US14. "Orphanet: Severe intellectual disability progressive spastic diplegia syndrome" (in en). https://www.orpha.net/consor/cgi-bin/OC_Exp.php?lng=EN&Expert=404473.
- ↑ "Urban's story – CTNNB1 Foundation" (in en-GB). https://ctnnb1-foundation.org/urbans-story/.
- ↑ "Severe intellectual disability-progressive spastic diplegia syndrome - About the Disease - Genetic and Rare Diseases Information Center" (in en). https://rarediseases.info.nih.gov/diseases/3505/severe-intellectual-disability-progressive-spastic-diplegia-syndrome.
- ↑ "Case Report: A de novo CTNNB1 Nonsense Mutation Associated With Neurodevelopmental Disorder, Retinal Detachment, Polydactyly" (in English). Frontiers in Pediatrics 8: 575673. 2020. doi:10.3389/fped.2020.575673. PMID 33425807.
- ↑ "Severe Intellectual Disability-Progressive Spastic Diplegia Syndrome" (in en). http://www.dovemed.com/diseases-conditions/severe-intellectual-disability-progressive-spastic-diplegia-syndrome/.
- ↑ "Severe intellectual disability-progressive spastic diplegia syndrome" (in en-US). https://www.rarehematologynews.com/rarediseases/severe-intellectual-disability-progressive-spastic-diplegia-syndrome/.
- ↑ "Deletion of CTNNB1 in inhibitory circuitry contributes to autism-associated behavioral defects". https://academic.oup.com/hmg/article/25/13/2738/2525756.
- ↑ "Germline Mutations in CTNNB1 Associated With Syndromic FEVR or Norrie Disease". Investigative Ophthalmology & Visual Science 60 (1): 93–97. January 2019. doi:10.1167/iovs.18-25142. PMID 30640974.
- ↑ "Identification of a novel splice mutation in CTNNB1 gene in a Chinese family with both severe intellectual disability and serious visual defects". Neurological Sciences 40 (8): 1701–1704. August 2019. doi:10.1007/s10072-019-03823-5. PMID 30929091.
- ↑ "CTNNB1 Neurodevelopmental Disorder". GeneReviews. Seattle (WA): University of Washington, Seattle. 1993. http://www.ncbi.nlm.nih.gov/books/NBK580527/. Retrieved 2022-09-05.
- ↑ "Entry - #615075 - NEURODEVELOPMENTAL DISORDER WITH SPASTIC DIPLEGIA AND VISUAL DEFECTS; NEDSDV - OMIM" (in en-us). https://omim.org/entry/615075.
- ↑ "CTNNB1 gene: MedlinePlus Genetics" (in en). https://medlineplus.gov/genetics/gene/ctnnb1/.
- ↑ "CTNNB1 Gene - GeneCards | CTNB1 Protein | CTNB1 Antibody". https://www.genecards.org/cgi-bin/carddisp.pl?gene=CTNNB1.
- ↑ "CTNNB1". https://www.sigmaaldrich.com/MX/es/genes/ctnnb1.
- ↑ "Intellectual Disability in Children" (in en). https://www.webmd.com/parenting/baby/child-intellectual-disability.
- ↑ "A clinical primer on intellectual disability". Translational Pediatrics 9 (Suppl 1): S23–S35. February 2020. doi:10.21037/tp.2020.02.02. PMID 32206581.
- ↑ "CTNNB1 syndrome – CTNNB1 Foundation" (in en-GB). https://ctnnb1-foundation.org/ctnnb1-syndrome/.
- ↑ "Diagnostic exome sequencing in persons with severe intellectual disability". The New England Journal of Medicine 367 (20): 1921–1929. November 2012. doi:10.1056/NEJMoa1206524. PMID 23033978.
- ↑ 26.0 26.1 "Dominant β-catenin mutations cause intellectual disability with recognizable syndromic features". The Journal of Clinical Investigation 124 (4): 1468–1482. April 2014. doi:10.1172/JCI70372. PMID 24614104.
- ↑ "A new intellectual disability syndrome caused by CTNNB1 haploinsufficiency". American Journal of Medical Genetics. Part A 164A (6): 1571–1575. June 2014. doi:10.1002/ajmg.a.36484. PMID 24668549.
- ↑ "De novo mutations in beta-catenin (CTNNB1) appear to be a frequent cause of intellectual disability: expanding the mutational and clinical spectrum". Human Genetics 134 (1): 97–109. January 2015. doi:10.1007/s00439-014-1498-1. PMID 25326669.
- ↑ "Clinical features associated with CTNNB1 de novo loss of function mutations in ten individuals". European Journal of Medical Genetics 60 (2): 130–135. February 2017. doi:10.1016/j.ejmg.2016.11.008. PMID 27915094.
- ↑ "Exome sequencing identifies a de novo mutation of CTNNB1 gene in a patient mainly presented with retinal detachment, lens and vitreous opacities, microcephaly, and developmental delay: Case report and literature review". Medicine 96 (20): e6914. May 2017. doi:10.1097/MD.0000000000006914. PMID 28514307.
- ↑ "Defects in the Cell Signaling Mediator β-Catenin Cause the Retinal Vascular Condition FEVR". American Journal of Human Genetics 100 (6): 960–968. June 2017. doi:10.1016/j.ajhg.2017.05.001. PMID 28575650.
- ↑ "CTNNB1 Foundation – CTNNB1 Syndrome" (in en-GB). https://ctnnb1-foundation.org/.
- ↑ "CTNNB1 Syndrome | Ctnnb1.org | About Us" (in en). https://www.ctnnb1.org/about_us.
- ↑ "Advancing CTNNB1 Cures & Treatments | Be Part of the Cure" (in en-US). https://www.curectnnb1.org/.
- ↑ "Paula nació con síndrome CTNNB1. Nuestra Unidad Infantil de Bilbao la ayuda a vivir mejor | Red Menni" (in es-ES). https://xn--daocerebral-2db.es/paula-nacio-con-sindrome-ctnnb1-nuestra-unidad-infantil-de-bilbao-la-ayuda-a-vivir-mejor/.
- ↑ "¿Qué es el síndrome CTNNB1, el trastorno que se confunde con el autismo?" (in es). 2021-05-26. https://www.mundodeportivo.com/vidae/salud/20210526/493941347676/sindrome-ctnnb1-trastorno-alteracion-cromosoma-autismo-act-pau.html.
- ↑ "Regusa solidaria - Dando a conocer el síndrome CTNNB1" (in es). 2022-05-05. https://gruporegusa.com/2022/05/05/regusa-solidaria-dando-a-conocer-el-sindrome-ctnnb1/.
- ↑ ""Si mi hija consigue andar y hablar sería algo grandioso"" (in es). 2021-07-12. https://www.elcorreo.com/bizkaia/nervion/hija-consigue-andar-20210713210131-nt.html.
- ↑ "Diez familias se unen para dar a conocer un síndrome que se confunde con el autismo" (in es-ES). 2021-05-23. https://www.20minutos.es/noticia/4705798/0/diez-familias-se-unen-para-dar-a-conocer-un-sindrome-que-se-confunde-con-el-autismo/.
 |

