Biology:SARS coronavirus
| Coronavirus | |
|---|---|

| |
| Virus classification | |
| Group: | Group IV ((+)ssRNA)
|
| Order: | |
| Family: | |
| Subfamily: | |
| Genus: | |
| Species: | SARS coronavirus
|
The SARS coronavirus, sometimes shortened to SARS-CoV, is the virus that causes severe acute respiratory syndrome (SARS).[1] On April 16, 2003, following the outbreak of SARS in Asia and secondary cases elsewhere in the world, the World Health Organization (WHO) issued a press release stating that the coronavirus identified by a number of laboratories was the official cause of SARS. Samples of the virus are being held in laboratories in New York City , San Francisco , Manila, Hong Kong, and Toronto.
On April 12, 2003, scientists working at the Michael Smith Genome Sciences Centre in Vancouver, British Columbia finished mapping the genetic sequence of a coronavirus believed to be linked to SARS. The team was led by Dr. Marco Marra and worked in collaboration with the British Columbia Centre for Disease Control and the National Microbiology Laboratory in Winnipeg, Manitoba, using samples from infected patients in Toronto. The map, hailed by the WHO as an important step forward in fighting SARS, is shared with scientists worldwide via the GSC website (see below). Dr. Donald Low of Mount Sinai Hospital in Toronto described the discovery as having been made with "unprecedented speed".[2] The sequence of the SARS coronavirus has since been confirmed by other independent groups.
The SARS coronavirus is one of several viruses identified by WHO as a likely cause of a future epidemic in a new plan developed after the Ebola epidemic for urgent research and development before and during an epidemic toward new diagnostic tests, vaccines and medicines.[3][4]
SARS
SARS,der per or Severe Acute Respiratory Syndrome, is the disease caused by SARS coronavirus. It causes an often severe illness marked initially by systemic symptoms of muscle pain, headache, and fever, followed in 2–14 days by the onset of respiratory symptoms,[5] mainly cough, dyspnea, and pneumonia. Another common finding in SARS patients is a decrease in the number of lymphocytes circulating in the blood.[6]
In the SARS outbreak of 2003, about 9% of patients with confirmed SARS infection died.[7] The mortality rate was much higher for those over 50 years old, with mortality rates approaching 50% for this subset of patients.[7]
History
The CDC and Canada's National Microbiology Laboratory identified the SARS genome in April, 2003.[8][9] Scientists at Erasmus University in Rotterdam, the Netherlands demonstrated that the SARS coronavirus fulfilled Koch's postulates thereby confirming it as the causative agent. In the experiments, macaques infected with the virus developed the same symptoms as human SARS victims.[10]
In late May 2003, studies from samples of wild animals sold as food in the local market in Guangdong, China, found the SARS coronavirus could be isolated from masked palm civets (Paguma sp.), but the animals did not always show clinical signs. The preliminary conclusion was the SARS virus crossed the xenographic barrier from palm civet to humans, and more than 10,000 masked palm civets were killed in Guangdong Province. Virus was also later found in raccoon dogs (Nyctereuteus sp.), ferret badgers (Melogale spp.), and domestic cats. In 2005, two studies identified a number of SARS-like coronaviruses in Chinese bats.[11][12] Phylogenetic analysis of these viruses indicated a high probability that SARS coronavirus originated in bats and spread to humans either directly or through animals held in Chinese markets. The bats did not show any visible signs of disease, but are the likely natural reservoirs of SARS-like coronaviruses. In late 2006, scientists from the Chinese Centre for Disease Control and Prevention of Hong Kong University and the Guangzhou Centre for Disease Control and Prevention established a genetic link between the SARS coronavirus appearing in civets and humans, confirming claims that the disease had jumped across species.[13]
Virology
The SARS coronavirus is a positive and single stranded RNA virus belonging to a family of enveloped coronaviruses. Its genome is about 29.7kb, which is one of the largest among RNA viruses. The SARS virus has 13 known genes and 14 known proteins. There are 265 nucleotides in the 5'UTR and 342 nucleotides in the 3'UTR. SARS is similar to other coronaviruses in that its genome expression starts with translation of two large ORFs, 1a and 1b, both of which are polyproteins.
The functions of several of these proteins are known:[14] ORFs 1a and 1b encode the replicase and there are four major structural proteins: nucleocapsid, spike, membrane and envelope. It also encodes for eight unique proteins, known as the accessory proteins, all with no known homologues. The function of these accessory proteins remains unknown.[15]
Coronaviruses usually express pp1a (the ORF1a polyprotein) and the PP1ab polyprotein with joins ORF1a and ORF1b. The polyproteins are then processed by enzymes that are encoded by ORF1a. Product proteins from the processing includes various replicative enzymes such as RNA dependent polymerase, RNA helicase, and proteinase. The replication complex in coronavirus is also responsible for the synthesis of various mRNAs downstream of ORF 1b, which are structural and accessory proteins. Two different proteins, 3CLpro and PL2pro, cleave the large polyproteins into 16 smaller subunits.
SARS-Coronavirus follows the replication strategy typical of the Coronavirus genus.
Morphology
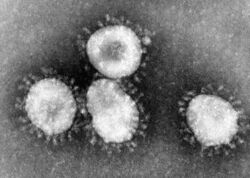
The morphology of the SARS coronavirus is characteristic of the coronavirus family as a whole. These viruses have large pleomorphic spherical particles with bulbous surface projections that form a corona around particles. The envelope of the virus contains lipid and appears to consist of a distinct pair of electron dense shells.
The internal component of the shell is a single-stranded helical ribonucleoprotein. There are also long surface projections that protrude from the lipid envelope. The size of these particles are about 80–90 nm.
Evolution
SARS is most closely related to group 2 coronaviruses, but it does not segregate into any of the other three groups of coronaviruses. A theory has been proposed that bat coronaviruses have been coevolved with their hosts for a long time then jumped species from bats to humans.[16][17] The closest outgroup to the coronaviruses are the toroviruses, with which it has homology in the ORF 1b replicase and the two viron proteins of S and M. SARS was determined to be an early split off from the group 2 coronaviruses based on a set of conserved domains that it shares with group 2.
A main difference between group 2 coronavirus and SARS is the nsp3 replicase subunit encoded by ORF1a. SARS does not have a papain-like proteinase 1.
Symptoms
Once a person has contracted SARS, the first symptom that they present with is a fever of at least 38 °C (100.4 °F) or higher. The early symptoms last about 2–7 days and include non-specific flu-like symptoms, including chills/rigor, muscle aches, headaches, diarrhea, sore throat, runny nose, malaise, and myalgia (muscle pain). Next, they develop a dry cough, shortness of breath, and an upper respiratory tract infection.
At that time, a chest x-ray is ordered to confirm pneumonia. If the chest appears clear and SARS is still suspected, a HRCT scan will be ordered, because it is visible earlier on this scan. In severe cases, it develops into respiratory failure and acute respiratory distress syndrome (ARDS), and in 70-90% of the cases, they develop lymphopenia (low count of lymphocyte white blood cells).
The incubation period for SARS-CoV is from 2–10 days, sometimes lasting up to 13 days, with a mean of 5 days.[5] Thus, symptoms usually develop between 2–10 days following infection by the virus. As part of the immune response, IgM antibody to the SARS-CoV is produced. This peaks during the acute or early convalescent phase (week 3) and declines by week 12. IgG antibody is produced later and peaks at week 12.[18]
Engineering the virus
Engineering of SARS virus has been done. In a paper published in 2006, a new transcription circuit was engineered to make recombinant SARS viruses. The recombination allowed for efficient expression of viral transcripts and proteins. The engineering of this transcription circuit reduces the RNA recombinant progeny viruses. The TRS (transcription regulatory sequences) circuit regulates efficient expression of SARS-CoV subgenomic mRNAs. The wild type TRS is ACGAAC.
A double mutation results in TRS-1 (ACGGAT) and a triple mutation results in TRS-2 (CCGGAT). When the remodeled TRS circuit containing viruses are genetically recombined with wild type TRS circuits, the result is a circuit reduced in production of subgenomic mRNA. The goal of modifying the SARS virus with this approach is to produce chimeric progeny that have reduced viability due to the incompatibility of the WT and engineered TRS circuits.
Novel subunit vaccine constructs for an S protein SARS vaccine based on the receptor binding domain (RBD) are being developed by the New York Blood Center. The re-emergence of SARS is possible, and the need remains for commercial vaccine and therapeutic development. However, the cost and length of time for product development, and the uncertain future demand, result in unfavorable economic conditions to accomplish this task. In the development of therapeutics and next-generation vaccines, more work is required to determine the structure/ function relationships of critical enzymes and structural proteins.
See also
- Carlo Urbani
- Progress of the SARS outbreak
- Severe acute respiratory syndrome
- SL-CoV-WIV1
Notes
- ↑ Thiel V (editor). (2007). Coronaviruses: Molecular and Cellular Biology (1st ed.). Caister Academic Press. ISBN 978-1-904455-16-5.
- ↑ "B.C. lab cracks suspected SARS code". CBCNews, Canada. April 2003. Archived from the original on 2007-11-26. https://web.archive.org/web/20071126060924/http://www.cbc.ca/canada/story/2003/04/12/sars_code030412.html.
- ↑ Kieny, Marie-Paule. "After Ebola, a Blueprint Emerges to Jump-Start R&D". Archived from the original on 20 December 2016. https://web.archive.org/web/20161220134725/https://blogs.scientificamerican.com/guest-blog/after-ebola-a-blueprint-emerges-to-jump-start-r-d/. Retrieved 13 December 2016.
- ↑ "LIST OF PATHOGENS". Archived from the original on 20 December 2016. https://web.archive.org/web/20161220180509/http://www.who.int/csr/research-and-development/list_of_pathogens/en/. Retrieved 13 December 2016.
- ↑ 5.0 5.1 Chan-Yeung M; Xu RH (November 2003). "SARS: epidemiology". Respirology (Carlton, Vic.) 8 (Suppl): S9–14. doi:10.1046/j.1440-1843.2003.00518.x. PMID 15018127.
- ↑ Yang M; Li CK; Li K; Hon KL; Ng MH; Chan PK; Fok TF (August 2004). "Hematological findings in SARS patients and possible mechanisms (review)". International Journal of Molecular Medicine 14 (2): 311–5. doi:10.3892/ijmm.14.2.311. PMID 15254784. Archived from the original on 2015-09-24. https://web.archive.org/web/20150924104246/http://www.spandidos-publications.com/ijmm/14/2/311.
- ↑ 7.0 7.1 Sørensen MD; Sørensen B; Gonzalez-Dosal R; Melchjorsen CJ; Weibel J; Wang J; Jun CW; Huanming Y et al. (May 2006). "Severe acute respiratory syndrome (SARS): development of diagnostics and antivirals". Annals of the New York Academy of Sciences 1067: 500–5. doi:10.1196/annals.1354.072. PMID 16804033.
- ↑ "Remembering SARS: A Deadly Puzzle and the Efforts to Solve It". Centers for Disease Control and Prevention. 11 April 2013. Archived from the original on 1 August 2013. https://web.archive.org/web/20130801143131/http://www.cdc.gov/about/history/sars/feature.htm. Retrieved 3 August 2013.
- ↑ "Coronavirus never before seen in humans is the cause of SARS". United Nations World Health Organization. 16 April 2006. Archived from the original on 12 August 2004. https://web.archive.org/web/20040812124322/http://www.who.int/mediacentre/releases/2003/pr31/en/. Retrieved 5 July 2006.
- ↑ "Aetiology: Koch's postulates fulfilled for SARS virus". Nature 423 (6937): 240. 2003. doi:10.1038/423240a. PMID 12748632.
- ↑ "Bats are natural reservoirs of SARS-like coronaviruses". Science 310 (5748): 676–9. 2005. doi:10.1126/science.1118391. PMID 16195424.
- ↑ "Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats". Proc. Natl. Acad. Sci. U.S.A. 102 (39): 14040–5. 2005. doi:10.1073/pnas.0506735102. PMID 16169905.
- ↑ "Scientists prove SARS-civet cat link". China Daily. 23 November 2006. Archived from the original on 14 June 2011. https://web.archive.org/web/20110614103319/http://www.chinadaily.com.cn/china/2006-11/23/content_740511.htm.
- ↑ McBride R; Fielding BC (November 2012). "The role of severe syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis". Viruses 4 (11): 2902–23. doi:10.3390/v4112902. PMID 23202509. PMC 3509677. Archived from the original on 2014-03-10. https://web.archive.org/web/20140310154558/http://www.mdpi.com/1999-4915/4/11/2902.
- ↑ McBride, Ruth; Fielding, Burtram C. (2012-11-07). "The Role of Severe Acute Respiratory Syndrome (SARS)-Coronavirus Accessory Proteins in Virus Pathogenesis". Viruses 4 (11): 2902–2923. doi:10.3390/v4112902. ISSN 1999-4915. PMID 23202509.
- ↑ Cui J; Han N; Streicker D; Li G; Tang X; Shi Z; Hu Z; Zhao G et al. (Oct 2007). "Evolutionary relationships between bat coronaviruses and their hosts". Emerg. Infect. Dis. 13 (10): 1526–32. doi:10.3201/eid1310.070448. PMID 18258002.
- ↑ Ge XY; Li JL; Yang XL; Chmura AA; Zhu G; Epstein JH; Mazet JK; Hu B et al. (Nov 28, 2013). "Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor". Nature 503 (7477): 535–8. doi:10.1038/nature12711. PMID 24172901.
- ↑ Bermingham A; Heinen P; Iturriza-Gómara M; Gray J; Appleton H; Zambon MC (July 2004). "Laboratory diagnosis of SARS". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 359 (1447): 1083–9. doi:10.1098/rstb.2004.1493. PMID 15306394.
References
- "Coronavirus as a possible cause of severe acute respiratory syndrome". Lancet 361 (9366): 1319–25. April 2003. doi:10.1016/s0140-6736(03)13077-2. PMID 12711465. http://linkinghub.elsevier.com/retrieve/pii/S0140673603130772.
- "Characterization of a Novel Coronavirus Associated with Severe Acute Respiratory Syndrome". Science 300 (5624): 1394–9. 30 May 2003. doi:10.1126/science.1085952. PMID 12730500. http://www.sciencemag.org/cgi/content/full/300/5624/1394.
- Marco A. Marra (30 May 2003). "The Genome Sequence of the SARS-Associated coronavirus". Science 300 (5624): 1399–1404. doi:10.1126/science.1085953. PMID 12730501. http://www.sciencemag.org/cgi/content/full/300/5624/1399.
- "Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage". J Mol Biol 331 (5): 991–1004. 29 August 2003. doi:10.1016/S0022-2836(03)00865-9. PMID 12927536.
- "Rewiring the severe acute respiratory syndrome coronavirus (SARS-CoV) transcription circuit: engineering a recombination-resistant genome". Proc Natl Acad Sci U S A 103 (33): 12546–51. 15 August 2006. doi:10.1073/pnas.0605438103. PMID 16891412. PMC 1531645. https://www.ncbi.nlm.nih.gov/sites/entrez.
- Thiel V, ed (2007). Coronaviruses: Molecular and Cellular Biology (1st ed.). Caister Academic Press. ISBN 978-1-904455-16-5.
- "Coronavirus Replication and Interaction with Host". Animal Viruses: Molecular Biology. Caister Academic Press. 2008. ISBN 978-1-904455-22-6.
External links
- WHO press release identifying and naming the SARS virus
- The SARS virus genetic map
- Science special on the SARS virus (free content: no registration required)
- U.S. Centers for Disease Control and Prevention (CDC) SARS home
- World Health Organization on alert
Wikidata ☰ Q278567 entry