Biology:MT-ND1
 Generic protein structure example |
MT-ND1 is a gene of the mitochondrial genome coding for the NADH-ubiquinone oxidoreductase chain 1 (ND1) protein.[1] The ND1 protein is a subunit of NADH dehydrogenase, which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain.[2] Variants of the human MT-ND1 gene are associated with mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS), Leigh's syndrome (LS), Leber's hereditary optic neuropathy (LHON) and increases in adult BMI.[3][4][5]
Structure
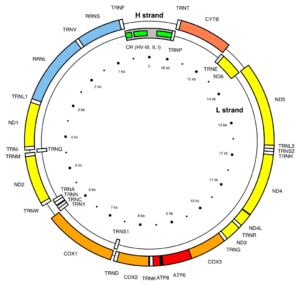
MT-ND1 is located in mitochondrial DNA from base pair 3,307 to 4,262.[1] The MT-ND1 gene produces a 36 kDa protein composed of 318 amino acids.[6][7] MT-ND1 is one of seven mitochondrial genes encoding subunits of the enzyme NADH dehydrogenase (ubiquinone), together with MT-ND2, MT-ND3, MT-ND4, MT-ND4L, MT-ND5, and MT-ND6. Also known as Complex I, this enzyme is the largest of the respiratory complexes. The structure is L-shaped with a long, hydrophobic transmembrane domain and a hydrophilic domain for the peripheral arm that includes all the known redox centres and the NADH binding site. The MT-ND1 product and the rest of the mitochondrially encoded subunits are the most hydrophobic of the subunits of Complex I and form the core of the transmembrane region.[2]
Function
MT-ND1-encoded NADH-ubiquinone oxidoreductase chain 1 is a subunit of the respiratory chain Complex I that is supposed to belong to the minimal assembly of core proteins required to catalyze NADH dehydrogenation and electron transfer to ubiquinone (coenzyme Q10).[8] Initially, NADH binds to Complex I and transfers two electrons to the isoalloxazine ring of the flavin mononucleotide (FMN) prosthetic arm to form FMNH2. The electrons are transferred through a series of iron-sulfur (Fe-S) clusters in the prosthetic arm and finally to coenzyme Q10 (CoQ), which is reduced to ubiquinol (CoQH2). The flow of electrons changes the redox state of the protein, resulting in a conformational change and pK shift of the ionizable side chain, which pumps four hydrogen ions out of the mitochondrial matrix.[2]
Clinical significance
Pathogenic variants of the mitochondrial gene MT-ND1 are known to cause mtDNA-associated Leigh syndrome, as are variants of MT-ATP6, MT-TL1, MT-TK, MT-TW, MT-TV, MT-ND2, MT-ND3, MT-ND4, MT-ND5, MT-ND6 and MT-CO3. Abnormalities in mitochondrial energy generation result in neurodegenerative disorders like Leigh syndrome, which is characterized by an onset of symptoms between 12 months and three years of age. The symptoms frequently present themselves following a viral infection and include movement disorders and peripheral neuropathy, as well as hypotonia, spasticity and cerebellar ataxia. Roughly half of affected individuals die of respiratory or cardiac failure by the age of three. Leigh syndrome is a maternally inherited disorder and its diagnosis is established through genetic testing of the aforementioned mitochondrial genes, including MT-ND1.[3] The m.4171C>A/MT-ND1 mutation also leads to a Leigh-like phenotype as well as bilateral brainstem lesions affecting the vestibular nuclei, resulting in vision loss, vomiting and vertigo.[4] These complex I genes have been associated with a variety of neurodegenerative disorders, including Leber's hereditary optic neuropathy (LHON), mitochondrial encephalomyopathy with stroke-like episodes (MELAS), overlap between LHON and MELAS,[9][10] and the previously mentioned Leigh syndrome.
Mitochondrial dysfunction resulting from variants of MT-ND1, MT-ND2 and MT-ND4L have been linked to BMI in adults and implicated in metabolic disorders including obesity, diabetes and hypertension.[5]
References
- ↑ 1.0 1.1 "Entrez Gene: MT-ND1 NADH dehydrogenase subunit 1". https://www.ncbi.nlm.nih.gov/gene/4535.
- ↑ 2.0 2.1 2.2 Voet, Donald J.; Voet, Judith G.; Pratt, Charlotte W. (2013). "Chapter 18, Mitochondrial ATP synthesis". Fundamentals of Biochemistry (4th ed.). Hoboken, NJ: Wiley. pp. 581–620. ISBN 978-0-47054784-7.
- ↑ 3.0 3.1 "Mitochondrial DNA-Associated Leigh Syndrome and NARP". GeneReviews [Internet]. Seattle (WA): University of Washington, Seattle. 1993–2015. https://www.ncbi.nlm.nih.gov/books/NBK1173.
- ↑ 4.0 4.1 "Association of the mtDNA m.4171C>A/MT-ND1 mutation with both optic neuropathy and bilateral brainstem lesions". BMC Neurology 14: 116. May 2014. doi:10.1186/1471-2377-14-116. PMID 24884847.
- ↑ 5.0 5.1 "Mitochondrial genetic variants identified to be associated with BMI in adults". PLOS ONE 9 (8): e105116. 2014. doi:10.1371/journal.pone.0105116. PMID 25153900. Bibcode: 2014PLoSO...9j5116F.
- ↑ "Integration of cardiac proteome biology and medicine by a specialized knowledgebase". Circulation Research 113 (9): 1043–53. October 2013. doi:10.1161/CIRCRESAHA.113.301151. PMID 23965338.
- ↑ "NADH-ubiquinone oxidoreductase chain 1". Cardiac Organellar Protein Atlas Knowledgebase (COPaKB). https://amino.heartproteome.org/web/protein/P03886.
- ↑ "MT-ND1 - NADH-ubiquinone oxidoreductase chain 1 - Homo sapiens (Human)". The UniProt Consortium. https://www.uniprot.org/uniprot/P03886.
- ↑ "A MELAS-associated ND1 mutation causing leber hereditary optic neuropathy and spastic dystonia". Archives of Neurology 64 (6): 890–3. June 2007. doi:10.1001/archneur.64.6.890. PMID 17562939.
- ↑ "LHON/MELAS overlap syndrome associated with a mitochondrial MTND1 gene mutation". European Journal of Human Genetics 13 (5): 623–7. May 2005. doi:10.1038/sj.ejhg.5201363. PMID 15657614.
Further reading
- "Harvesting the fruit of the human mtDNA tree". Trends in Genetics 22 (6): 339–45. June 2006. doi:10.1016/j.tig.2006.04.001. PMID 16678300.
- "Dinucleotide repeat in the human mitochondrial D-loop". Human Molecular Genetics 1 (2): 140. May 1992. doi:10.1093/hmg/1.2.140-a. PMID 1301157.
- "Differentiation of HT-29 human colonic adenocarcinoma cells correlates with increased expression of mitochondrial RNA: effects of trehalose on cell growth and maturation". Cancer Research 52 (13): 3718–25. July 1992. PMID 1377597.
- "An ND-6 mitochondrial DNA mutation associated with Leber hereditary optic neuropathy". Biochemical and Biophysical Research Communications 187 (3): 1551–7. September 1992. doi:10.1016/0006-291X(92)90479-5. PMID 1417830.
- "A new mtDNA mutation associated with Leber hereditary optic neuroretinopathy". American Journal of Human Genetics 48 (6): 1147–53. June 1991. PMID 1674640.
- "Normal variants of human mitochondrial DNA and translation products: the building of a reference data base". Human Genetics 88 (2): 139–45. December 1991. doi:10.1007/bf00206061. PMID 1757091.
- "Alternative, simultaneous complex I mitochondrial DNA mutations in Leber's hereditary optic neuropathy". Biochemical and Biophysical Research Communications 174 (3): 1324–30. February 1991. doi:10.1016/0006-291X(91)91567-V. PMID 1900003.
- "Leber hereditary optic neuropathy: identification of the same mitochondrial ND1 mutation in six pedigrees". American Journal of Human Genetics 49 (5): 939–50. November 1991. PMID 1928099.
- "Electron transfer properties of NADH:ubiquinone reductase in the ND1/3460 and the ND4/11778 mutations of the Leber hereditary optic neuroretinopathy (LHON)". FEBS Letters 292 (1–2): 289–92. November 1991. doi:10.1016/0014-5793(91)80886-8. PMID 1959619.
- "Replication-competent human mitochondrial DNA lacking the heavy-strand promoter region". Molecular and Cellular Biology 11 (3): 1631–7. March 1991. doi:10.1128/MCB.11.3.1631. PMID 1996112.
- "Leber hereditary optic neuropathy: involvement of the mitochondrial ND1 gene and evidence for an intragenic suppressor mutation". American Journal of Human Genetics 48 (5): 935–42. May 1991. PMID 2018041.
- "Seven unidentified reading frames of human mitochondrial DNA encode subunits of the respiratory chain NADH dehydrogenase". Cold Spring Harbor Symposia on Quantitative Biology 51 (1): 103–14. 1987. doi:10.1101/sqb.1986.051.01.013. PMID 3472707.
- "URF6, last unidentified reading frame of human mtDNA, codes for an NADH dehydrogenase subunit". Science 234 (4776): 614–8. October 1986. doi:10.1126/science.3764430. PMID 3764430. Bibcode: 1986Sci...234..614C.
- "Six unidentified reading frames of human mitochondrial DNA encode components of the respiratory-chain NADH dehydrogenase". Nature 314 (6012): 592–7. 1985. doi:10.1038/314592a0. PMID 3921850. Bibcode: 1985Natur.314..592C.
- "Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing". Journal of Molecular Biology 143 (2): 161–78. October 1980. doi:10.1016/0022-2836(80)90196-5. PMID 6260957.
- "Interaction of rhodanese with mitochondrial NADH dehydrogenase". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology 742 (2): 278–84. January 1983. doi:10.1016/0167-4838(83)90312-6. PMID 6402020.
- "Sequence and organization of the human mitochondrial genome". Nature 290 (5806): 457–65. April 1981. doi:10.1038/290457a0. PMID 7219534. Bibcode: 1981Natur.290..457A.
- "Distinctive features of the 5'-terminal sequences of the human mitochondrial mRNAs". Nature 290 (5806): 465–70. April 1981. doi:10.1038/290465a0. PMID 7219535. Bibcode: 1981Natur.290..465M.
- "Recent African origin of modern humans revealed by complete sequences of hominoid mitochondrial DNAs". Proceedings of the National Academy of Sciences of the United States of America 92 (2): 532–6. January 1995. doi:10.1073/pnas.92.2.532. PMID 7530363. Bibcode: 1995PNAS...92..532H.
- "A new mitochondrial DNA mutation associated with non-insulin-dependent diabetes mellitus". Biochemical and Biophysical Research Communications 209 (2): 664–8. April 1995. doi:10.1006/bbrc.1995.1550. PMID 7733935.
- "Whole-exome sequencing identifies a variant of the mitochondrial MT-ND1 gene associated with epileptic encephalopathy: west syndrome evolving to Lennox-Gastaut syndrome". Human Mutation 34 (12): 1623–7. December 2013. doi:10.1002/humu.22445. PMID 24105702.
- "Intragenic inversion of mtDNA: a new type of pathogenic mutation in a patient with mitochondrial myopathy". American Journal of Human Genetics 66 (6): 1900–4. June 2000. doi:10.1086/302927. PMID 10775530.
External links
 |